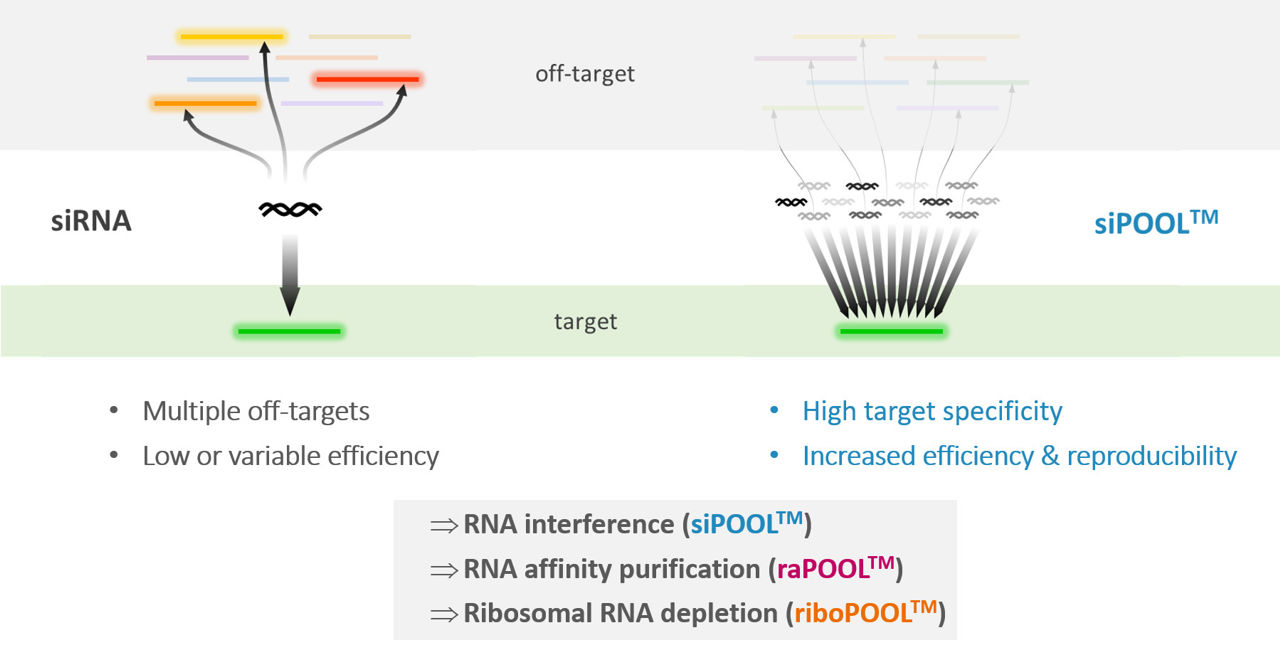
Why do we pool siRNAs?
At siTOOLs Biotech, we develop complex pools of selected oligos.
Oligo complexity guarantees robust and high efficacy by cooperative activity of multiple probes of one target.
The low concentration of each oligo dilutes undesired side effects and yields high reagent specificity.
siPOOLs are a set of 30 selected siRNAs with diverse seed sequences to minimize off-target effects and improve silencing robustness (Hannus et al., 2014).
Obtain reliable results with one single pool avoiding redunant experiments with multiple single siRNAs.
Learn more about siPOOLs
riboPOOLs are highly complex sets of biotinylated probes. rRNA pulldown is strongly enhanced by multiple probes binding to each rRNA fragment.
Pan-riboPOOLs cover groups of species with proprietary combination of probes targeting conserved ribosomal regions.
rRNA depletion of strongly fragemented RNA samples is achieved by a probe set with maximum probe density.
Learn more about riboPOOLs
raPOOLs are custom designed complex probe sets to enrich RNAs of interest.
Probe complexity allows reliable pull down of native RNAs with their interacting partners.
For Capture-Seq requires affordable, highly complex probeset for multiple genes.
Learn more about raPOOLs
Request a quote
Order now