| rRNA depletion for any species | High complexity probes for efficient, reliable rRNA removal | Cost-effective |
| Wide RNA-input range (10 ng – 3 µg) | riboPOOLs for highly degraded samples (FFPE) | special solution for ribosome profiling |

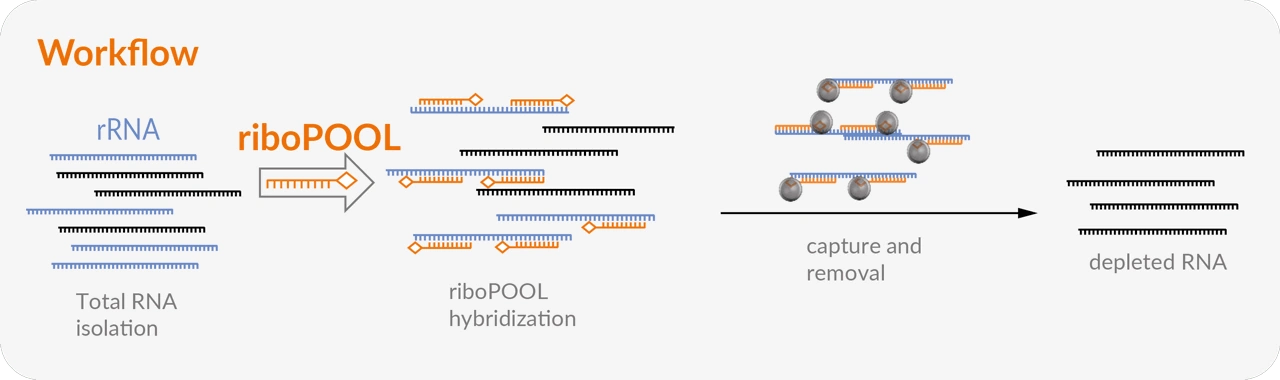
The Pan-riboPOOLs are a versatile rRNA depletion solution that allows for simple mono- and multitranscriptomic studies using a single-step rRNA depletion for a phylogenetic group (e.g., bacteria, fungi, or mammals).
The option to combine riboPOOLs facilitates single-step rRNA depletion in mixed samples such as environmental, blood, or infected tissue (e.g., SARS-CoV-2) samples. The so-called combination riboPOOLs enable simple metatranscriptomics studies by combining 2-4 riboPOOLs of non-related species.
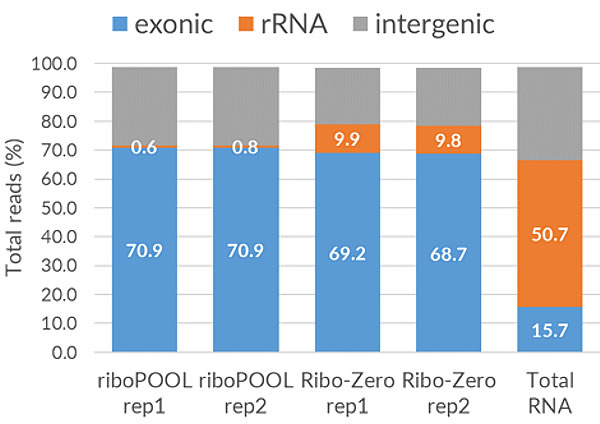
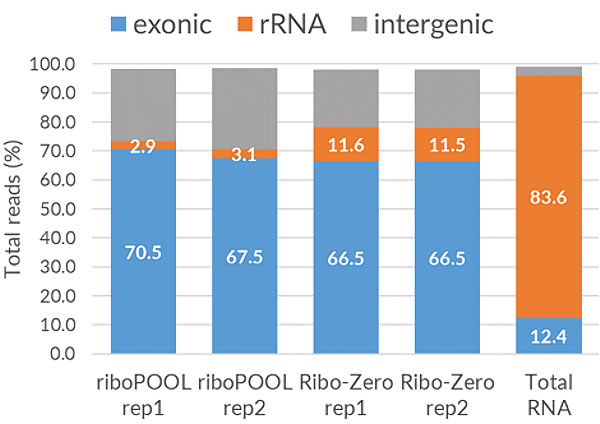
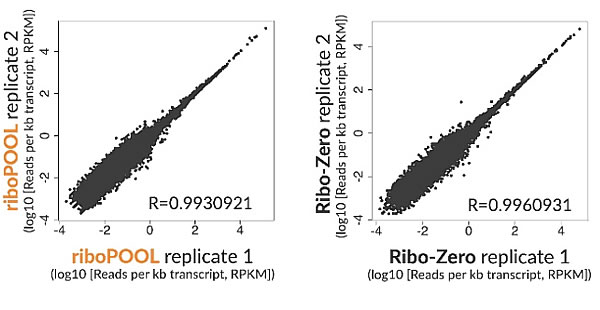
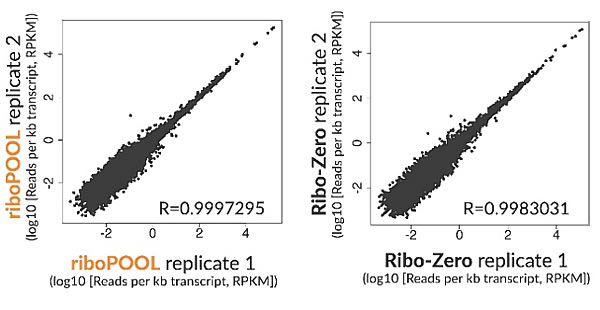
Single-species riboPOOLs are available for well-studied and lesser-known species (Escherichia coli, Arabidopsis thaliana or Schmidtea mediterranea and many more). Single-species riboPOOLs are specifically designed based on the species' rRNA to target both conserved and non-conserved regions.
Moreover, single species riboPOOLs are used on high quality to medium quality RNA and result in high rRNA depletion efficiency.
|
Bacteria Escherichia coli Bacillus subtilis Caulobacter crescentus Clostridium perfringens Stenotrophomonas sp. Salmonella enterica Pseudomonas aeruginosa Archaea Haloferax volcanii Plants Oryza sativa Arabidopsis thaliana Azolla filiculoides Vertebrata Mus musculus / R. norvegicus Homo sapiens sapiens Chinchilla lanigera Gallus gallus domesticus Felis catus NEW Equus caballus NEW Porifera Amphimedon queenslandica Fungi Saccharomyces cerevisiae Filamentous-Fungi Pichia pastoris Staphylococcus aureus Ustilago maydis Schizosaccharomyces pombe |
Arthropoda Nematoda: |
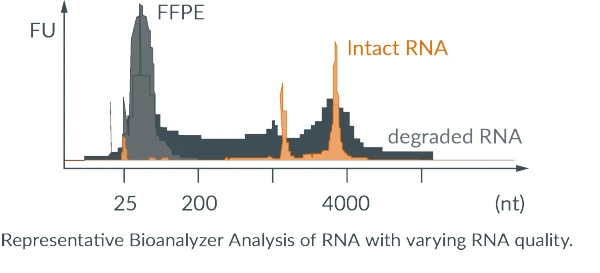
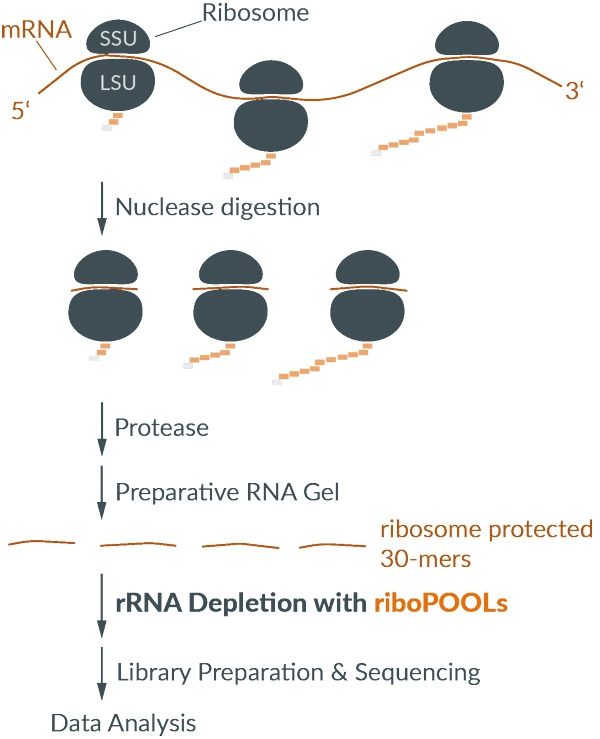
FFPE /degraded | Ribo-Seq
|
Is your species not listed here? Let us set up a Custom riboPOOL for your specific species!
Custom riboPOOLs include a one-time setup fee. You can purchase the custom riboPOOL as a ready-made riboPOOL at any kit or probe size.
Please use the form below to request for a quote or ask for more information about riboPOOLs.